Hi, I am Samuel Botter Martins (Samuka) Resume in pdf
I am a senior data science and an adjunct professor of Computer Science and Data Science with more than 7 years of experience. I have a theoretical background and practical experience designing innovative machine learning solutions for various applications, such as computer vision, medical image analysis, and Cheminformatics. In recent years, I have successfully supervised over 20 students on research data science projects and published impactful work in relevant conferences and journals.
My excellent communication skills, cultivated as a radio broadcaster, teacher, and YouTube creator (tech channel with 300k+ views), enable me to simplify complex concepts and effectively engage diverse audiences. I am driven to make a positive impact by designing cutting-edge data science solutions and am excited to explore new opportunities and collaborations.
Let's connect and see how I can bring my experience, skills, and unique perspective to your organization.
My research interests are:
- machine learning
- deep learning
- (medical) image processing and analysis
- computer vision
- visual analytics
Experience
-
09/2023 – present
Senior Data Scientist
Itaú Unibanco - São Paulo-SP (Brazil)
- Developing propensity models and time series anomaly detection methods.
- Technologies: Python, AWS, Scikit-Learn, and Pandas.
-
08/2016 – present
YouTube Creator
Channel: xavecoding (aimed at Portuguese speakers)
- Over 300 videos, 6,000 subscribers, and 320,000 views.
- Channel dedicated to courses and tutorials on ML and computer science topics.
-
10/2020 – 09/2023
Coordinator of the Data Science Specialization
Federal Institute of Education, Science, and Technology of São Paulo (IFSP) - Campinas-SP (Brazil)
- Coordinated the curriculum committee responsible for curriculum development and educational policy.
- Managed faculty and conducted admission processes with over 300 candidates.
- Organized a local data science workshop with 100+ participants.
- Strived to ensure that the program offers a comprehensive education in data science and equips students with the skills they need to thrive in this rapidly evolving field.
-
07/2016 – 09/2023
Adjunct Professor of Data Science and Computer Science
Federal Institute of Education, Science, and Technology of São Paulo (IFSP) - Campinas-SP (Brazil)
- Conducted classes for undergraduate and graduate students, such as Applied Statistics, Machine Learning, Deep Learning, and Natural Language Processing.
- Guided and mentored 20+ graduate and undergraduate students in data science research projects.
- Developed and published novel machine-learning solutions for problems such as computer vision, medical image analysis, and Cheminformatics.
- Technologies: C, Python, Java, Keras, OpenCV, Scikit-Learn, and Pandas.
-
01/2012 – 12/2012
Web Developer
Tray E-Commerce Platform, Marília-SP, Brazil
Developed four modules for an e-commerce platform on Ruby Rails and MySQL.
-
10/2006 – 02/2016
Radio Broadcaster (volunteer work)
Millenium FM 104.9, Pompéia-SP, Brazil
- Hosted talk shows and other radio programs.
- Produced audio commercials and radio spots.
- Served as master of ceremonies at corporate and public events.
Education
-
03/2015 – 11/2020
Ph.D. in Machine Learning (double degree)
University of Campinas (Brazil) & University of Groningen (Netherlands)
Research on Machine Learning for Medical Image Analysis. Ph.D. thesis
- Published 14 research papers on machine learning and received 3 awards at scientific congresses
- Designed automatic unsupervised solutions to detect brain anomalies in MR images.
- Combination of image processing (e.g., superpixels) and one-class classification (OC-SVM).
- High anomaly detection rates (86%+) on stroke images with a reduction by up 20x false positives.
- Developed a deep-learning-based approach to detect abnormal hippocampi from epilepsy patients.
- Detection accuracies from 86% to 100% (in some specific scenarios).
- Applied visual analytics to understand the model and results, improving accuracy by up 13%.
- Proposed an automatic method based on statistical learning (probabilistic models and texture classifications) for anomalous brain image segmentation - reduced segmentation errors by up 15%.
- Technologies: C, Python, OpenCV, Scikit-Learn, Pandas, ITKSNAP.
-
03/2013 – 02/2015
M.Sc. in Machine Learning
University of Campinas (Brazil)
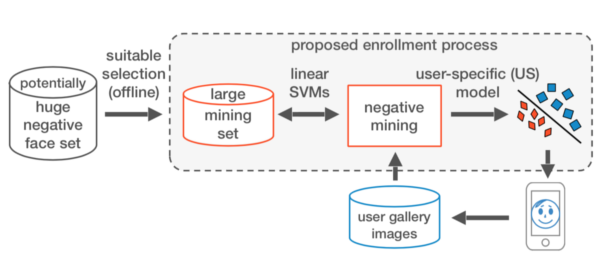
Research on Machine Learning for Face Recognition and Negative Mining. Dissertation
- Investigated state-of-the-art deep features for face recognition in unconstrained scenarios.
- Designed an SVM-based method that mines informative negative samples within interactive times.
- Technologies: C, Python, OpenCV, TensorFlow, Scikit-Learn.
-
03/2008 – 12/2012
B.Sc. in Computer Science
University of São Paulo (Brazil)
Expertise
Machine Learning and Deep Learning
Several supervised, unsupervised, and weakly-supervised learning algorithms applied to problems of regression, clustering, and classification.
Structured and unstructured data (images).
Convolutional Neural Networks, GAN, transfer learning.
Experiment design and quantitative analysis.
(Medical) Image Processing and Analysis
An wide range of traditional automatic and interactive techniques for image preprocessing, segmentation, and characterization (feature extraction).
Automatic and semi-automatic segmentation and detection of anomalies in medical images (e.g., brain MR images, chest CT images).
Computer Vision
Experience on different image applications: face recognition in the wild, object detection, and semantic segmentation.
Data Visualization
Different techniques for data analysis.
Visual analytics methods to understand neural networks.
Data Engineering with AWS
Creation and management of Data Warehouses, Data Lakes, and Lakehouses in AWS
Development of ETL/ELT
PySpark, AWS Redshift, AWS Glue, AWS Athena
Skills
- Python, C/C++, Java, SQL
- HTML, CSS, Javascript, React, D3.js
- Git/GitHub, Linux
- ITK snap, napari
- AWS
- Matplotlib, Seaborn, Plotly
- Scikit-learn, XGboost, PyCaret
- Keras, Pytorch
- Scikit-image, OpenCV, nibabel
- SpaCy, NLTK
- English (fluent)
- Italian (basic)
- Portuguese (native)
- Scientific and paper writing.
- Mentoring (under)graduate students in research projects.
- Scientific mindset.
- Strong communication skills.
Selected Papers
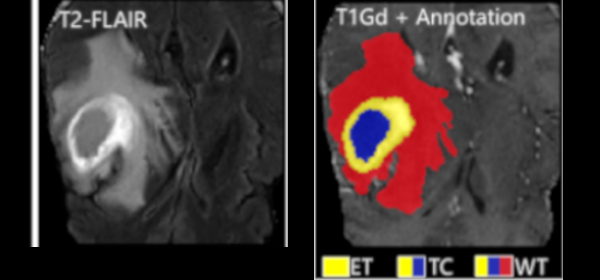
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Sarthak Pati, Samuel B. Martins, et al.
Nature Communications 13, 7346 (2022)
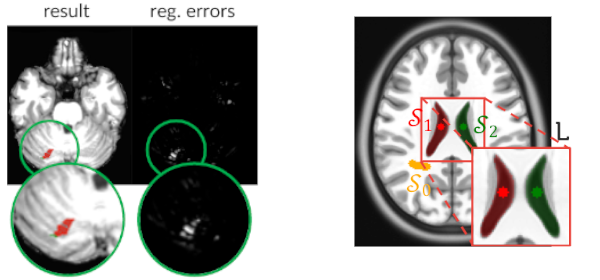
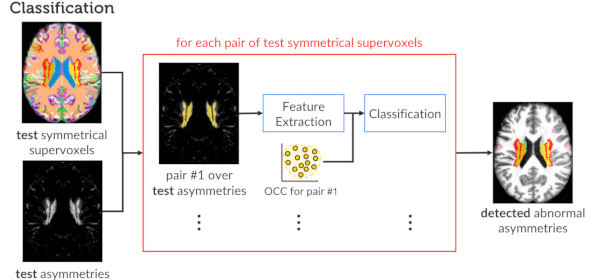
Published versionCombining Registration Errors and Supervoxel Classification for Unsupervised Brain Anomaly Detection
Samuel B. Martins, Alexandre X. Falcão, Alexandru C. Telea
Biomedical Engineering Systems and Technologies, Springer International Publishing, pp. 140–164, 2021.
Published versionInvestigating the impact of supervoxel segmentation for unsupervised abnormal brain asymmetry detection
Samuel B. Martins, Alexandru C. Telea, Alexandre X. Falcão
Computerized Medical Imaging and Graphics, 85, pp. 101770, 2020.
Published versionAn adaptive probabilistic atlas for anomalous brain segmentation in MR images
Samuel B. Martins, Jordão Bragantini, Alexandre X. Falcão, Clarissa L. Yasuda
Medical Physics, 46 (11), pp. 4940-4950, 2019.
Published versionMethods: In AdaPro, we first build one probabilistic atlas per object of interest from a training set with normal 3D images and the corresponding 3D object masks. Second, we incorporate a texture classifier based on convex optimization which dynamically indicates the regions of the target 3D image where the PAs (shape constraints) should be further adapted. This strategy is mathematically more elegant and avoids problems with post-processing. Third, we add a new object-based delineation algorithm based on combinatorial optimization and diffusion filtering. AdaPro can then be used to locate and delineate the objects in the coordinate space of the atlas or of the test image. We also compare AdaPro with three other state-of-the-art methods: an statistical shape model based on synergistic object search and delineation, and two methods based on multi-atlas label fusion (MALF).
Results: We evaluate the methods quantitatively on 3D MR-T1 brain images of 2T and 3T from epilepsy patients, before and after temporal lobe resections, and on the template and native coordinate spaces. The results show that AdaPro is considerably faster and consistently more accurate than the baselines with statistical significance in both coordinate spaces.
Conclusion: AdaPro can be used as a fast and effective step for brain tissue segmentation and it can also be easily extended to segment subcortical brain structures. By choice of its components, probabilistic atlas, texture classifier, and delineation algorithm, it can also be extended to other organs and imaging modalities.
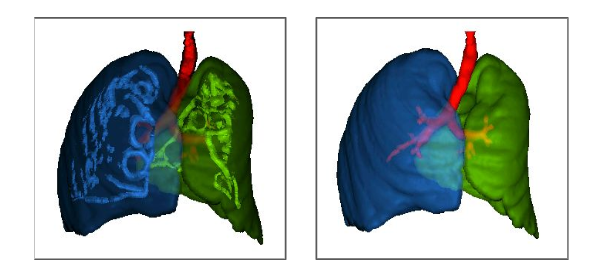
ALTIS: A fast and automatic lung and trachea CT-image segmentation method
Azael M. Sousa, Samuel B. Martins, Alexandre X. Falcão, Fabiano Reis, Ericson Bagatin, Klaus Irion
Medical Physics, 46 (11), pp. 4970-4982., 2019.
Published versionMethods: ALTIS consists of a sequence of image foresting transforms (IFTs) organized in three main steps: (a) lung-and-trachea extraction, (b) seed estimation inside background, trachea, left lung, and right lung, and (c) their delineation such that each object is defined by an optimum-path forest rooted at its internal seeds. We compare ALTIS with two methods based on shape models (SOSM-S and MALF), and one algorithm based on seeded region growing (PTK).
Results: The experiments involve the highest number of scans found in literature - 1255 scans, from multiple public data sets containing many anomalous cases, being only 50 normal scans used for training and 1205 scans used for testing the methods. Quantitative experiments are based on two metrics, DICE and ASSD. Furthermore, we also demonstrate the robustness of ALTIS in seed estimation. Considering the test set, the proposed method achieves an average DICE of 0.987 for both lungs and 0.898 for the trachea, whereas an average ASSD of 0.938 for the right lung, 0.856 for the left lung, and 1.316 for the trachea. These results indicate that ALTIS is statistically more accurate and considerably faster than the compared methods, being able to complete segmentation in a few seconds on modern PCs.
Conclusion: ALTIS is the most effective and efficient choice among the compared methods to segment left lung, right lung, and trachea in anomalous CT scans for subsequent detection, segmentation, and quantitative analysis of abnormal structures in the lung parenchyma and pleural space.
A Fast and Robust Negative Mining Approach for Enrollment in Face Recognition Systems
Samuel B. Martins, Giovani Chiachia, Alexandre X. Falcão
Conference on Graphics, Patterns and Images (SIBGRAPI), IEEE, pp. 201-208, 2017.
Published versionAwards
- Best Ph.D. Thesis Award of the Workshop of Theses and Dissertations (WTD) 2021
- Conference on Graphics, Patterns and Images (SIBGRAPI),
- Thesis: Unsupervised Brain Anomaly Detection in MR Images.
- Best Student Paper Awards 2020
- BIOSTECT BIOIMAGING, Valleta, Malta
- Paper: BADRESC: Brain Anomaly Detection based on Registration Errors and Supervoxel Classification.
- Best Student Paper Award Finalist 2017
- SPIE Medical Imaging, Orlando, USA
- Paper: A Multi-Object Statistical Atlas Adaptive for Deformable Registration Errors in Anomalous Medical Image Segmentation.
Talks
- Panel discussion: Exchange and International Experience Dec 2022
- XVII Workshop of Theses, Dissertations and Scientific Initiation Works
- Online, UNICAMP, Brazil
- Cultura do Aprender: Data Science & Analytics Dec 2022
- Culture of Learning: Data Science and Analytics
- Meet Up CPFL Energia
- Online
- Primeiros passos em Ciência de Dados Nov 2022
- First steps in Data Science
- 19th National Science and Technology Week of Federal Institute of São Paulo
- Hortolândia-SP, Brazil
- Inteligência Artificial para a Análise de Imagens Médicas Oct 2020
- Artificial Intelligence for Image Analysis Medical
- 7th National Science and Technology Week of Federal Institute of São Paulo
- Online
Registered Software
- SaMI - Plataforma Inteligente Voltada à Saúde Materno Infantil
- Intelligent Platform for Maternal and Child Health.
Other Interests
- Technology
- Reading
- Board games
- Games
- Piano, Guitar
- Soccer, Volleyball
- Miniature painting, Toy makeover
Feel free to send me an e-mail or get in touch on my social media by clicking the badges below to see my profile.